Reference¶
Module functions¶
This package implements the louvain algorithm in C++ and exposes it to
python. It relies on (python-)igraph for it to function. Besides the
relative flexibility of the implementation, it also scales well, and can be run
on graphs of millions of nodes (as long as they can fit in memory). Each method
is represented by a different class, all of whom derive from
MutableVertexPartition. In addition,
multiplex graphs are supported as layers, which also supports multislice
representations.
Examples
The simplest example just finds a partition using modularity
>>> G = ig.Graph.Tree(100, 3)
>>> partition = louvain.find_partition(G, louvain.ModularityVertexPartition)
Alternatively, one can access the different optimisation routines individually
and construct partitions oneself. These partitions can then be optimised by
constructing an Optimiser object and running
optimise_partition().
>>> G = ig.Graph.Tree(100, 3)
>>> partition = louvain.CPMVertexPartition(G, resolution_parameter = 0.1)
>>> optimiser = louvain.Optimiser()
>>> diff = optimiser.optimise_partition(partition)
The Optimiser class contains also the different subroutines that are
used internally by optimise_partition(). In addition, through
the Optimiser class there are various options available for changing some of
the optimisation procedure which can affect both speed and quality, which are
not immediately available in louvain.find_partition().
- louvain.find_partition(graph, partition_type, initial_membership=None, weights=None, seed=None, **kwargs)¶
Detect communities using the default settings.
This function detects communities given the specified method in the
partition_type. This should be type derived fromVertexPartition.MutableVertexPartition, e.g.ModularityVertexPartitionorCPMVertexPartition. Optionally an initial membership and edge weights can be provided. Remaining**kwargsare passed on to the constructor of thepartition_type, including for example aresolution_parameter.- Parameters
graph (
ig.Graph) – The graph for which to detect communities.partition_type (type of :class:`) – The type of partition to use for optimisation.
initial_membership (list of int) – Initial membership for the partition. If
Nonethen defaults to a singleton partition.weights (list of double, or edge attribute) – Weights of edges. Can be either an iterable or an edge attribute.
seed (int) – Seed for the random number generator. By default uses a random seed if nothing is specified.
**kwargs – Remaining keyword arguments, passed on to constructor of
partition_type.
- Returns
The optimised partition.
- Return type
partition
See also
Examples
>>> G = ig.Graph.Famous('Zachary') >>> partition = louvain.find_partition(G, louvain.ModularityVertexPartition)
- louvain.find_partition_multiplex(graphs, partition_type, seed=None, **kwargs)¶
Detect communities for multiplex graphs.
Each graph should be defined on the same set of vertices, only the edges may differ for different graphs. See
Optimiser.optimise_partition_multiplex()for a more detailed explanation.- Parameters
graphs (list of
ig.Graph) – List oflouvain.VertexPartitionlayers to optimise.partition_type (type of
MutableVertexPartition) – The type of partition to use for optimisation (identical for all graphs).seed (int) – Seed for the random number generator. By default uses a random seed if nothing is specified.
**kwargs – Remaining keyword arguments, passed on to constructor of
partition_type.
- Returns
list of int – membership of nodes.
float – Improvement in quality of combined partitions, see
Optimiser.optimise_partition_multiplex().
Notes
We don’t return a partition in this case because a partition is always defined on a single graph. We therefore simply return the membership (which is the same for all layers).
Examples
>>> n = 100 >>> G_1 = ig.Graph.Lattice([n], 1) >>> G_2 = ig.Graph.Lattice([n], 1) >>> membership, improvement = louvain.find_partition_multiplex([G_1, G_2], ... louvain.ModularityVertexPartition)
- louvain.find_partition_temporal(graphs, partition_type, interslice_weight=1, slice_attr='slice', vertex_id_attr='id', edge_type_attr='type', weight_attr='weight', seed=None, **kwargs)¶
Detect communities for temporal graphs.
Each graph is considered to represent a time slice and does not necessarily need to be defined on the same set of vertices. Nodes in two consecutive slices are identified on the basis of the
vertex_id_attr, i.e. if two nodes in two consecutive slices have an identical value of thevertex_id_attrthey are coupled. Thevertex_id_attrshould hence be unique in each slice. The nodes are then coupled with a weight ofinterslice_weightwhich is set in the edge attributeweight_attr. No weight is set if theinterslice_weightis None (i.e. corresponding in practice with a weight of 1). Seetime_slices_to_layers()for a more detailed explanation.- Parameters
graphs (list of
ig.Graph) – List oflouvain.VertexPartitionlayers to optimise.partition_type (type of
VertexPartition.MutableVertexPartition) – The type of partition to use for optimisation (identical for all graphs).interslice_weight (float) – The weight of the coupling between two consecutive time slices.
slice_attr (string) – The vertex attribute to use for indicating the slice of a node.
vertex_id_attr (string) – The vertex to use to identify nodes.
edge_type_attr (string) – The edge attribute to use for indicating the type of link (interslice or intraslice).
weight_attr (string) – The edge attribute used to indicate the weight.
seed (int) – Seed for the random number generator. By default uses a random seed if nothing is specified.
**kwargs – Remaining keyword arguments, passed on to constructor of
partition_type.
- Returns
list of membership – list containing for each slice the membership vector.
float – Improvement in quality of combined partitions, see
Optimiser.optimise_partition_multiplex().
See also
Examples
>>> n = 100 >>> G_1 = ig.Graph.Lattice([n], 1) >>> G_1.vs['id'] = range(n) >>> G_2 = ig.Graph.Lattice([n], 1) >>> G_2.vs['id'] = range(n) >>> membership, improvement = louvain.find_partition_temporal([G_1, G_2], ... louvain.ModularityVertexPartition, ... interslice_weight=1)
- louvain.slices_to_layers(G_coupling, slice_attr='slice', vertex_id_attr='id', edge_type_attr='type', weight_attr='weight')¶
Convert a coupling graph of slices to layers of graphs.
This function converts a graph of slices to layers so that they can be used with this package. This function assumes that the slices are represented by nodes in
G_coupling, and stored in the attributeslice_attr. In other words,G_coupling.vs[slice_attr]should containig.Graphs . The slices will be converted to layers, and nodes in different slices will be coupled if the two slices are connected inG_coupling. Nodes in two connected slices are identified on the basis of thevertex_id_attr, i.e. if two nodes in two connected slices have an identical value of thevertex_id_attrthey will be coupled. Thevertex_id_attrshould hence be unique in each slice. The weight of the coupling is determined by the weight of this link inG_coupling, as determined by theweight_attr.- Parameters
G_coupling (
ig.Graph) – The graph connecting the different slices.slice_attr (string) – The vertex attribute which contains the slices.
vertex_id_attr (string) – The vertex attribute which is used to identify whether two nodes in two slices represent the same node, and hence, should be coupled.
edge_type_attr (string) – The edge attribute to use for indicating the type of link (
intersliceorintraslice).weight_attr (string) – The edge attribute used to indicate the (coupling) weight.
- Returns
G_layers (list of
ig.Graph) – A list of slices converted to layers.G_interslice (
ig.Graph) – The interslice coupling layer.G (
ig.Graph) – The complete graph containing all layers and interslice couplings.
Notes
The distinction between slices and layers is not easy to grasp. Slices in this context refer to graphs that somehow represents different aspects of a network. The simplest example is probably slices that represents time: there are different snapshots network across time, and each snapshot is considered a slice. Some nodes may drop out of the network over time, while others enter the network. Edges may change over time, or the weight of the links may change over time. This is just the simplest example of a slice, and there may be different, more complex possibilities. Below an example with three time slices:
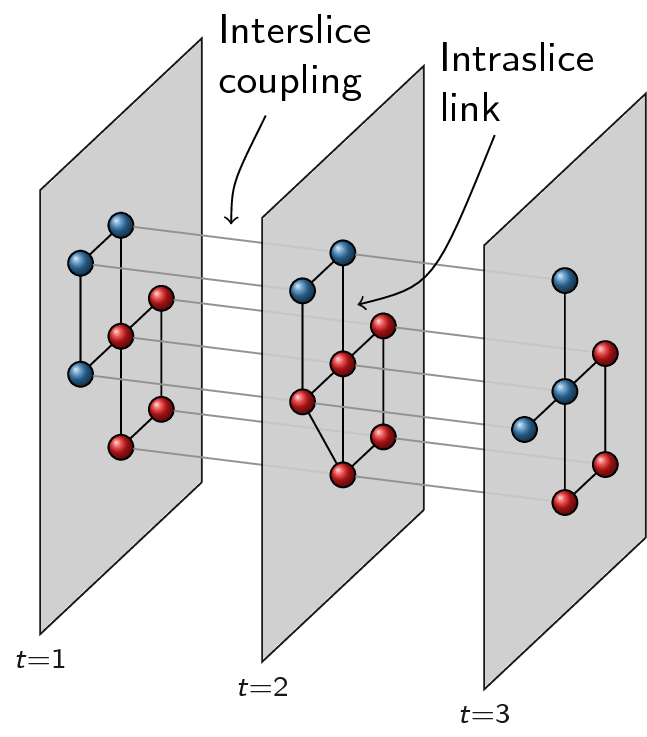
Now in order to optimise partitions across these different slices, we represent them slightly differently, namely as layers. The idea of layers is that all graphs always are defined on the same set of nodes, and that only the links differ for different layers. We thus create new nodes as combinations of original nodes and slices. For example, if node 1 existed in both slice 1 and in slice 2, we will thus create two nodes to build the layers: a node 1-1 and a node 1-2. Additionally, if the slices are connected in the slice graph, the two nodes would also be connected, so there would be a linke between node 1-1 and 1-2. Different slices will then correspond to different layers: each layer only contains the link for that particular slice. In addition, for methods such as
CPMVertexPartition, so-callednode_sizesare required, and for them to properly function, they should be set to 0 (which is handled appropriately in this function, and stored in the vertex attributenode_size). We thus obtain equally many layers as we have slices, and we need one more layer for representing the interslice couplings. For the example provided above, we thus obtain the following: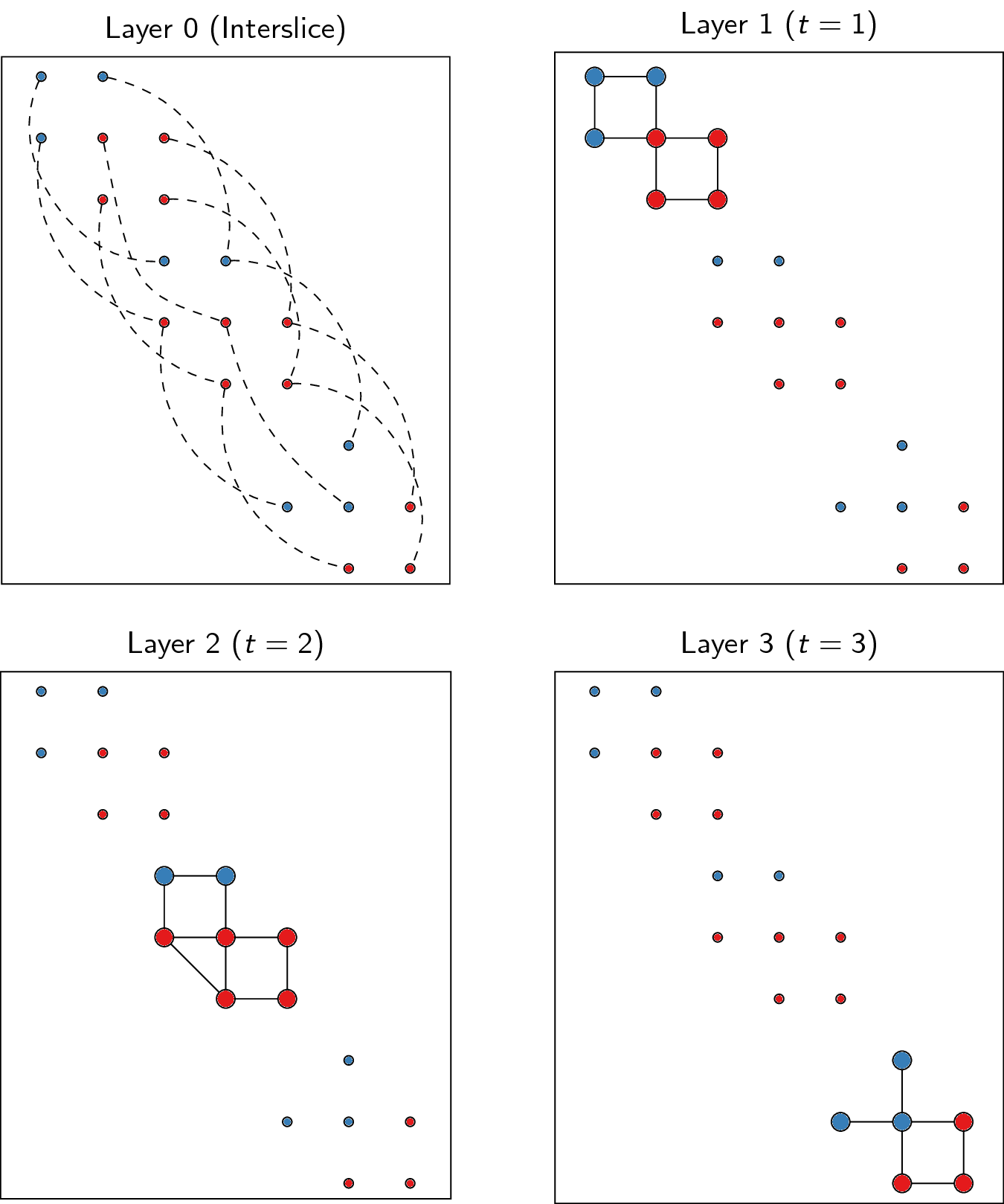
The idea of doing community detection with slices is further detailed in [1].
References
- 1
Mucha, P. J., Richardson, T., Macon, K., Porter, M. A., & Onnela, J.-P. (2010). Community structure in time-dependent, multiscale, and multiplex networks. Science, 328(5980), 876-8. 10.1126/science.1184819
- louvain.time_slices_to_layers(graphs, interslice_weight=1, slice_attr='slice', vertex_id_attr='id', edge_type_attr='type', weight_attr='weight')¶
Convert time slices to layer graphs.
Each graph is considered to represent a time slice. This function simply connects all the consecutive slices (i.e. the slice graph) with an
interslice_weight. The further conversion is then delegated toslices_to_layers(), which also provides further details.See also
Optimiser¶
- class louvain.Optimiser¶
Bases:
objectClass for doing community detection using the Louvain algorithm.
The optimiser class provides a number of different methods for optimising a given partition. The overall optimise procedure
optimise_partition()callsmove_nodes()then aggregates the graph and repeats the same procedure. For calculating the actual improvement of moving a node (corresponding a subset of nodes in the aggregate graph), the code relies ondiff_move()which provides different values for different methods (e.g. modularity or CPM). Finally, the Optimiser class provides a routine to construct aresolution_profile()on a resolution parameter.Create a new Optimiser object
- property consider_comms¶
Determine how alternative communities are considered for moving a node for optimising a partition.
Nodes will only move to alternative communities that improve the given quality function.
Notes
This attribute should be set to one of the following values
louvain.ALL_NEIGH_COMMSConsider all neighbouring communities for moving.louvain.ALL_COMMSConsider all communities for moving. This is especially useful in the case of negative links, in which case it may be better to move a node to a non-neighbouring community.louvain.RAND_NEIGH_COMMConsider a random neighbour community for moving. The probability to choose a community is proportional to the number of neighbours a node has in that community.louvain.RAND_COMMConsider a random community for moving. The probability to choose a community is proportional to the number of nodes in that community.
- property consider_empty_community¶
if
Trueconsider also moving nodes to an empty community (default).- Type
boolean
- move_nodes(partition, consider_comms=None)¶
Move nodes to alternative communities for optimising the partition.
- Parameters
partition – The partition for which to move nodes.
consider_comms – If
Noneusesconsider_comms, but can be set to something else.
- Returns
Improvement in quality function.
- Return type
float
Notes
When moving nodes, the function loops over nodes and considers moving the node to an alternative community. Which community depends on
consider_comms. The function terminates when no more nodes can be moved to an alternative community.Examples
>>> G = ig.Graph.Famous('Zachary') >>> optimiser = louvain.Optimiser() >>> partition = louvain.ModularityVertexPartition(G) >>> diff = optimiser.move_nodes(partition)
- optimise_partition(partition)¶
Optimise the given partition.
- Parameters
partition – The
MutableVertexPartitionto optimise.- Returns
Improvement in quality function.
- Return type
float
Examples
>>> G = ig.Graph.Famous('Zachary') >>> optimiser = louvain.Optimiser() >>> partition = louvain.ModularityVertexPartition(G) >>> diff = optimiser.optimise_partition(partition)
- optimise_partition_multiplex(partitions, layer_weights=None)¶
Optimise the given partitions simultaneously.
- Parameters
partitions – List of
MutableVertexPartitionlayers to optimise.layer_weights – List of weights of layers.
- Returns
Improvement in quality of combined partitions, see Notes.
- Return type
float
Notes
This method assumes that the partitions are defined for graphs with the same vertices. The connections between the vertices may be different, but the vertices themselves should be identical. In other words, all vertices should have identical indices in all graphs (i.e. node i is assumed to be the same node in all graphs). The quality of the overall partition is simply the sum of the individual qualities for the various partitions, weighted by the layer_weight. If we denote by \(Q_k\) the quality of layer \(k\) and the weight by \(\lambda_k\), the overall quality is then
\[Q = \sum_k \lambda_k Q_k.\]This is particularly useful for graphs containing negative links. When separating the graph in two graphs, the one containing only the positive links, and the other only the negative link, by supplying a negative weight to the latter layer, we try to find relatively many positive links within a community and relatively many negative links between communities. Note that in this case it may be better to assign a node to a community to which it is not connected so that
consider_commsmay be better set tolouvain.ALL_COMMS.Besides multiplex graphs where each node is assumed to have a single community, it is also useful in the case of for example multiple time slices, or in situations where nodes can have different communities in different slices. The package includes some special helper functions for using
optimise_partition_multiplex()in such cases, where there is a conversion required from (time) slices to layers suitable for use in this function.See also
slices_to_layers(),time_slices_to_layers(),find_partition_multiplex(),find_partition_temporal()Examples
>>> G_pos = ig.Graph.SBM(100, pref_matrix=[[0.5, 0.1], [0.1, 0.5]], block_sizes=[50, 50]) >>> G_neg = ig.Graph.SBM(100, pref_matrix=[[0.1, 0.5], [0.5, 0.1]], block_sizes=[50, 50]) >>> optimiser = louvain.Optimiser() >>> partition_pos = louvain.ModularityVertexPartition(G_pos) >>> partition_neg = louvain.ModularityVertexPartition(G_neg) >>> diff = optimiser.optimise_partition_multiplex( ... partitions=[partition_pos, partition_neg], ... layer_weights=[1,-1])
- resolution_profile(graph, partition_type, resolution_range, weights=None, bisect_func=<function Optimiser.<lambda>>, min_diff_bisect_value=1, min_diff_resolution=0.001, linear_bisection=False, number_iterations=1, **kwargs)¶
Use bisectioning on the resolution parameter in order to construct a resolution profile.
- Parameters
graph – The graph for which to construct a resolution profile.
partition_type – The type of
MutableVertexPartitionused to find a partition (must support resolution parameters obviously).resolution_range – The range of resolution values that we would like to scan.
weights – If provided, indicates the edge attribute to use as a weight.
bisect_func – The function used for bisectioning. For the methods currently implemented, this should usually not be altered.
min_diff_bisect_value – The difference in the value returned by the bisect_func below which the bisectioning stops (i.e. by default, a difference of a single edge does not trigger further bisectioning).
min_diff_resolution – The difference in resolution below which the bisectioning stops. For positive differences, the logarithmic difference is used by default, i.e.
diff = log(res_1) - log(res_2) = log(res_1/res_2), for whichdiff > min_diff_resolutionto continue bisectioning. Set the linear_bisection to true in order to use only linear bisectioning (in the case of negative resolution parameters for example, which can happen with negative weights).linear_bisection – Whether the bisectioning will be done on a linear or on a logarithmic basis (if possible).
number_iterations – Indicates the number of iterations of the algorithm to run. If negative (or zero) the algorithm is run until a stable iteration.
- Returns
A list of partitions for different resolutions.
- Return type
list of
MutableVertexPartition
Examples
>>> G = ig.Graph.Famous('Zachary') >>> optimiser = louvain.Optimiser() >>> profile = optimiser.resolution_profile(G, louvain.CPMVertexPartition, ... resolution_range=(0,1))
- set_rng_seed(value)¶
Set the random seed for the random number generator.
- Parameters
value – The integer seed used in the random number generator
MutableVertexPartition¶
- class louvain.VertexPartition.MutableVertexPartition(graph, initial_membership=None)¶
Bases:
VertexClusteringContains a partition of graph, derives from
ig.VertexClustering.This class contains the basic implementation for optimising a partition. Specifically, it implements all the administration necessary to keep track of the partition from various points of view. Internally, it keeps track of the number of internal edges (or total weight), the size of the communities, the total incoming degree (or weight) for a community, et cetera.
In order to keep the administration up-to-date, all changes in a partition should be done through
move_node()orset_membership(). The first moves a node from one community to another, and updates the administration. The latter simply updates the membership vector and updates the administration.The basic idea is that
diff_move()computes the difference in the quality function if we would callmove_node()for the same move. These functions are overridden in any derived classes to provide an actual implementation. These functions are used byOptimiserto optimise the partition.Warning
This base class should never be used in practice, since only derived classes provide an actual implementation.
- Parameters
graph – The ig.Graph on which this partition is defined.
membership – The membership vector of this partition. Membership[i] = c implies that node i is in community c. If None, it is initialised with a singleton partition community, i.e. membership[i] = i.
- classmethod FromPartition(partition, **kwargs)¶
Create a new partition from an existing partition.
- Parameters
partition – The
MutableVertexPartitionto replicate.**kwargs – Any remaining keyword arguments will be passed on to the constructor of the new partition.
Notes
This may for example come in handy when determining the quality of a partition using a different method. Suppose that we already have a partition
pand that we want to determine the Significance of that partition. We can then simply use>>> p = louvain.find_partition(ig.Graph.Famous('Zachary'), ... louvain.ModularityVertexPartition) >>> sig = louvain.SignificanceVertexPartition.FromPartition(p).quality()
- aggregate_partition(membership_partition=None)¶
Aggregate the graph according to the current partition and provide a default partition for it.
The aggregated graph can then be found as a parameter of the partition
partition.graph.Notes
This function contrasts to the function
cluster_graphin igraph itself, which also provides the aggregate graph, but we may require setting the appropriateresolution_parameter,weightsandnode_sizes. In particular, this function also ensures that the quality defined on the aggregate partition is identical to the quality defined on the original partition.Examples
>>> G = ig.Graph.Famous('Zachary') >>> partition = louvain.find_partition(G, louvain.ModularityVertexPartition) >>> aggregate_partition = partition.aggregate_partition(partition) >>> aggregate_graph = aggregate_partition.graph >>> aggregate_partition.quality() == partition.quality() True
- diff_move(v, new_comm)¶
Calculate the difference in the quality function if node
vis moved to communitynew_comm.- Parameters
v – The node to move.
new_comm – The community to move to.
- Returns
Difference in quality function.
- Return type
float
Notes
The difference returned by diff_move should be equivalent to first determining the quality of the partition, then calling move_node, and then determining again the quality of the partition and looking at the difference. In other words
>>> partition = louvain.find_partition(ig.Graph.Famous('Zachary'), ... louvain.ModularityVertexPartition) >>> diff = partition.diff_move(v=0, new_comm=0) >>> q1 = partition.quality() >>> partition.move_node(v=0, new_comm=0) >>> q2 = partition.quality() >>> round(diff, 10) == round(q2 - q1, 10) True
Warning
Only derived classes provide actual implementations, the base class provides no implementation for this function.
- from_coarse_partition(partition, coarse_node=None)¶
Update current partition according to coarser partition.
- Parameters
partition (
MutableVertexPartition) – The coarser partition used to update the current partition.coarse_node (list of int) – The coarser node which represent the current node in the partition.
Notes
This function is to be used to determine the correct partition for an aggregated graph. In particular, suppose we move nodes and then get an aggregate graph.
>>> diff = optimiser.move_nodes(partition) >>> aggregate_partition = partition.aggregate_partition()
Now we also move nodes in the aggregate partition
>>> diff = optimiser.move_nodes(aggregate_partition)
Now we improved the quality function of
aggregate_partition, but this is not yet reflected in the originalpartition. We can thus call>>> partition.from_coarse_partition(aggregate_partition)
so that
partitionnow reflects the changes made toaggregate_partition.The
coarse_nodecan be used it theaggregate_partitionis not defined based on the membership of this partition. In particular the membership of this partition is defined as follows:>>> for v in G.vs: ... partition.membership[v] = aggregate_partition.membership[coarse_node[v]]
If
coarse_nodeisNoneit is assumed the coarse node was defined based on the membership of the current partition, so that>>> for v in G.vs: ... partition.membership[v] = aggregate_partition.membership[partition.membership[v]]
This can be useful when the aggregate partition is defined on a more refined partition.
- move_node(v, new_comm)¶
Move node
vto communitynew_comm.- Parameters
v – Node to move.
new_comm – Community to move to.
Examples
>>> G = ig.Graph.Famous('Zachary') >>> partition = louvain.ModularityVertexPartition(G) >>> partition.move_node(0, 1)
- quality()¶
The current quality of the partition.
- renumber_communities()¶
Renumber the communities so that they are numbered in decreasing size.
Notes
The sort is not necessarily stable.
- set_membership(membership)¶
Set membership.
- total_possible_edges_in_all_comms()¶
The total possible number of edges in all communities.
Notes
If we denote by \(n_c\) the number of nodes in community \(c\), this is simply
\[\sum_c \binom{n_c}{2}\]
- total_weight_from_comm(comm)¶
The total weight (i.e. number of edges) from a community.
- Parameters
comm – Community
Notes
This includes all edges, also the ones that are internal to a community. Sometimes this is also referred to as the community (out)degree.
See also
total_weight_to_comm(),total_weight_in_comm(),total_weight_in_all_comms()
- total_weight_in_all_comms()¶
The total weight (i.e. number of edges) within all communities.
Notes
This should be equal to simply the sum of
total_weight_in_commfor all communities.See also
total_weight_to_comm(),total_weight_from_comm(),total_weight_in_comm()
- total_weight_in_comm(comm)¶
The total weight (i.e. number of edges) within a community.
- Parameters
comm – Community
See also
total_weight_to_comm(),total_weight_from_comm(),total_weight_in_all_comms()
- total_weight_to_comm(comm)¶
The total weight (i.e. number of edges) to a community.
- Parameters
comm – Community
Notes
This includes all edges, also the ones that are internal to a community. Sometimes this is also referred to as the community (in)degree.
See also
total_weight_from_comm(),total_weight_in_comm(),total_weight_in_all_comms()
- weight_from_comm(v, comm)¶
The total number of edges (or sum of weights) to node
vfrom communitycomm.See also
weight_to_comm()
- weight_to_comm(v, comm)¶
The total number of edges (or sum of weights) from node
vto communitycomm.See also
weight_from_comm()
ModularityVertexPartition¶
- class louvain.ModularityVertexPartition(graph, initial_membership=None, weights=None)¶
Bases:
MutableVertexPartitionImplements modularity.
Notes
The quality function is
\[Q = \frac{1}{2m} \sum_{ij} \left(A_{ij} - \frac{k_i k_j}{2m} \right)\delta(\sigma_i, \sigma_j)\]where \(A\) is the adjacency matrix, \(k_i\) is the degree of node \(i\), \(m\) is the total number of edges, \(\sigma_i\) denotes the community of node \(i\) and \(\delta(\sigma_i, \sigma_j) = 1\) if \(\sigma_i = \sigma_j\) and 0 otherwise.
This can alternatively be formulated as a sum over communities:
\[Q = \frac{1}{2m} \sum_{c} \left(m_c - \frac{K_c^2}{4m} \right)\]where \(m_c\) is the number of internal edges of community \(c\) and \(K_c = \sum_{i \mid \sigma_i = c} k_i\) is the total degree of nodes in community \(c\).
References
- 1
Newman, M. E. J., & Girvan, M. (2004). Finding and evaluating community structure in networks. Physical Review E, 69(2), 026113. 10.1103/PhysRevE.69.026113
- Parameters
graph (
ig.Graph) – Graph to define the partition on.initial_membership (list of int) – Initial membership for the partition. If
Nonethen defaults to a singleton partition.weights (list of double, or edge attribute) – Weights of edges. Can be either an iterable or an edge attribute.
RBConfigurationVertexPartition¶
- class louvain.RBConfigurationVertexPartition(graph, initial_membership=None, weights=None, resolution_parameter=1.0)¶
Bases:
LinearResolutionParameterVertexPartitionImplements Reichardt and Bornholdt’s Potts model with a configuration null model.
This quality function uses a linear resolution parameter.
Notes
The quality function is
\[Q = \sum_{ij} \left(A_{ij} - \gamma \frac{k_i k_j}{2m} \right)\delta(\sigma_i, \sigma_j)\]where \(A\) is the adjacency matrix, \(k_i\) is the degree of node \(i\), \(m\) is the total number of edges, \(\sigma_i\) denotes the community of node \(i\), \(\delta(\sigma_i, \sigma_j) = 1\) if \(\sigma_i = \sigma_j\) and 0 otherwise, and, finally \(\gamma\) is a resolution parameter.
This can alternatively be formulated as a sum over communities:
\[Q = \sum_{c} \left(m_c - \gamma\frac{K_c^2}{4m} \right)\]where \(m_c\) is the number of internal edges of community \(c\) and \(K_c = \sum_{i \mid \sigma_i = c} k_i\) is the total degree of nodes in community \(c\).
Note that this is the same as
ModularityVertexPartitionfor \(\gamma=1\) and using the normalisation by \(2m\).References
- 1
Reichardt, J., & Bornholdt, S. (2006). Statistical mechanics of community detection. Physical Review E, 74(1), 016110. 10.1103/PhysRevE.74.016110
- Parameters
graph (
ig.Graph) – Graph to define the partition on.initial_membership (list of int) – Initial membership for the partition. If
Nonethen defaults to a singleton partition.weights (list of double, or edge attribute) – Weights of edges. Can be either an iterable or an edge attribute.
resolution_parameter (double) – Resolution parameter.
- property resolution_parameter¶
Resolution parameter.
RBERVertexPartition¶
- class louvain.RBERVertexPartition(graph, initial_membership=None, weights=None, node_sizes=None, resolution_parameter=1.0)¶
Bases:
LinearResolutionParameterVertexPartitionImplements Reichardt and Bornholdt’s Potts model with a configuration null model.
This quality function uses a linear resolution parameter.
Notes
The quality function is
\[Q = \sum_{ij} \left(A_{ij} - \gamma p \right)\delta(\sigma_i, \sigma_j)\]where \(A\) is the adjacency matrix,
\[p = \frac{m}{\binom{n}{2}}\]is the overall density of the graph, \(\sigma_i\) denotes the community of node \(i\), \(\delta(\sigma_i, \sigma_j) = 1\) if \(\sigma_i = \sigma_j\) and 0 otherwise, and, finally \(\gamma\) is a resolution parameter.
This can alternatively be formulated as a sum over communities:
\[Q = \sum_{c} \left[m_c - \gamma p \binom{n_c}{2} \right]\]where \(m_c\) is the number of internal edges of community \(c\) and \(n_c\) the number of nodes in community \(c\).
References
- 1
Reichardt, J., & Bornholdt, S. (2006). Statistical mechanics of community detection. Physical Review E, 74(1), 016110. 10.1103/PhysRevE.74.016110
- Parameters
graph (
ig.Graph) – Graph to define the partition on.initial_membership (list of int) – Initial membership for the partition. If
Nonethen defaults to a singleton partition.weights (list of double, or edge attribute) – Weights of edges. Can be either an iterable or an edge attribute.
node_sizes (list of int, or vertex attribute) – Sizes of nodes are necessary to know the size of communities in aggregate graphs. Usually this is set to 1 for all nodes, but in specific cases this could be changed.
resolution_parameter (double) – Resolution parameter.
CPMVertexPartition¶
- class louvain.CPMVertexPartition(graph, initial_membership=None, weights=None, node_sizes=None, resolution_parameter=1.0)¶
Bases:
LinearResolutionParameterVertexPartitionImplements CPM. This quality function uses a linear resolution parameter.
Notes
The quality function is
\[Q = \sum_{ij} \left(A_{ij} - \gamma \right)\delta(\sigma_i, \sigma_j)\]where \(A\) is the adjacency matrix, \(\sigma_i\) denotes the community of node \(i\), \(\delta(\sigma_i, \sigma_j) = 1\) if \(\sigma_i = \sigma_j\) and 0 otherwise, and, finally \(\gamma\) is a resolution parameter.
This can alternatively be formulated as a sum over communities:
\[Q = \sum_{c} \left[m_c - \gamma \binom{n_c}{2} \right]\]where \(m_c\) is the number of internal edges of community \(c\) and \(n_c\) the number of nodes in community \(c\).
The resolution parameter \(\gamma\) for this functions has a particularly simple interpretation. The internal density of communities
\[p_c = \frac{m_c}{\binom{n_c}{2}} \geq \gamma\]is higher than \(\gamma\), while the external density
\[p_{cd} = \frac{m_{cd}}{n_c n_d} \leq \gamma\]is lower than \(\gamma\). In other words, choosing a particular \(\gamma\) corresponds to choosing to find communities of a particular density, and as such defines communities. Finally, the definition of a community is in a sense independent of the actual graph, which is not the case for any of the other methods (see the reference for more detail).
References
- 1
Traag, V. A., Van Dooren, P., & Nesterov, Y. (2011). Narrow scope for resolution-limit-free community detection. Physical Review E, 84(1), 016114. 10.1103/PhysRevE.84.016114
- Parameters
graph (
ig.Graph) – Graph to define the partition on.initial_membership (list of int) – Initial membership for the partition. If
Nonethen defaults to a singleton partition.weights (list of double, or edge attribute) – Weights of edges. Can be either an iterable or an edge attribute.
node_sizes (list of int, or vertex attribute) – Sizes of nodes are necessary to know the size of communities in aggregate graphs. Usually this is set to 1 for all nodes, but in specific cases this could be changed.
resolution_parameter (double) – Resolution parameter.
- Bipartite(resolution_parameter_01, resolution_parameter_0=0, resolution_parameter_1=0, degree_as_node_size=False, types='type', **kwargs)¶
Create three layers for bipartite partitions.
This creates three layers for bipartite partition necessary for detecting communities in bipartite networks. These three layers should be passed to
Optimiser.optimise_partition_multiplex()withlayer_weights=[1,-1,-1].- Parameters
graph (
ig.Graph) – Graph to define the bipartite partitions on.resolution_parameter_01 (double) – Resolution parameter for in between two classes.
resolution_parameter_0 (double) – Resolution parameter for class 0.
resolution_parameter_1 (double) – Resolution parameter for class 1.
degree_as_node_size (boolean) –
If
Trueuse degree as node size instead of 1, to mimic modularity, see Notes.types (vertex attribute or list) – Indicator of the class for each vertex. If not 0, 1, it is automatically converted.
**kwargs – Additional arguments passed on to default constructor of
CPMVertexPartition._notes-bipartite (..) –
Notes
For bipartite networks, we would like to be able to set three different resolution parameters: one for within each class \(\gamma_0, \gamma_1\), and one for the links between classes, \(\gamma_{01}\). Then the formulation would be
\[Q = \sum_{ij} [A_{ij} - (\gamma_0\delta(s_i,0) + \gamma_1\delta(s_i,1)) \delta(s_i,s_j) - \gamma_{01}(1 - \delta(s_i, s_j)) ]\delta(\sigma_i, \sigma_j)\]In terms of communities this is
\[Q = \sum_c (e_c - \gamma_{01} 2 n_c(0) n_c(1) - \gamma_0 n^2_c(0) - \gamma_1 n^2_c(1))\]where \(n_c(0)\) is the number of nodes in community \(c\) of class 0 (and similarly for 1) and \(e_c\) is the number of edges within community \(c\). We denote by \(n_c = n_c(0) + n_c(1)\) the total number of nodes in community \(c\).
We achieve this by creating three layers : (1) all nodes have
node_size = 1and all relevant links; (2) only nodes of class 0 havenode_size = 1and no links; (3) only nodes of class 1 havenode_size = 1and no links. If we add the first with resolution parameter \(\gamma_{01}\), and the others with resolution parameters \(\gamma_{01} - \gamma_0\) and \(\gamma_{01} - \gamma_1\), but the latter two with a layer weight of -1 while the first layer has layer weight 1, we obtain the following:\[\begin{split}Q &= \sum_c (e_c - \gamma_{01} n_c^2) -\sum_c (- (\gamma_{01} - \gamma_0) n_c(0)^2) -\sum_c (- (\gamma_{01} - \gamma_1) n_c(1)^2) \\ &= \sum_c [e_c - \gamma_{01} 2 n_c(0) n_c(1) - \gamma_{01} n_c(0)^2 - \gamma_{01} n_c(1)^2) + ( \gamma_{01} - \gamma_0) n_c(0)^2 + ( \gamma_{01} - \gamma_1) n_c(1)^2 ] \\ &= \sum_c [e_c - \gamma_{01} 2 n_c(0) n_c(1) - \gamma_{0} n_c(0)^2 - \gamma_{1} n_c(1)^2]\end{split}\]Although the derivation above is using \(n_c^2\), implicitly assuming a direct graph with self-loops, similar derivations can be made for undirected graphs using \(\binom{n_c}{2}\), but the notation is then somewhat more convoluted.
If we set node sizes equal to the degree, we get something similar to modularity, except that the resolution parameter should still be divided by \(2m\). In particular, in general (i.e. not specifically for bipartite graph) if
node_sizes=G.degree()we then obtain\[Q = \sum_{ij} A_{ij} - \gamma k_i k_j\]In the case of bipartite graphs something similar is obtained, but then correctly adapted (as long as the resolution parameter is also appropriately rescaled).
Note
This function is not suited for directed graphs in the case of using the degree as node sizes.
SignificanceVertexPartition¶
- class louvain.SignificanceVertexPartition(graph, initial_membership=None, node_sizes=None)¶
Bases:
MutableVertexPartitionImplements Significance.
Notes
The quality function is
\[Q = \sum_c \binom{n_c}{2} D(p_c \parallel p)\]where \(n_c\) is the number of nodes in community \(c\),
\[p_c = \frac{m_c}{\binom{n_c}{2}},\]is the density of community \(c\),
\[p = \frac{m}{\binom{n}{2}}\]is the overall density of the graph, and finally
\[D(x \parallel y) = x \ln \frac{x}{y} + (1 - x) \ln \frac{1 - x}{1 - y}\]is the binary Kullback-Leibler divergence.
For directed graphs simply multiply the binomials by 2. The expected Significance in Erdos-Renyi graphs behaves roughly as \(\frac{1}{2} n \ln n\) for both directed and undirected graphs in this formulation.
Warning
This method is not suitable for weighted graphs.
References
- 1
Traag, V. A., Krings, G., & Van Dooren, P. (2013). Significant scales in community structure. Scientific Reports, 3, 2930. 10.1038/srep02930
- Parameters
graph (
ig.Graph) – Graph to define the partition on.initial_membership (list of int) – Initial membership for the partition. If
Nonethen defaults to a singleton partition.node_sizes (list of int, or vertex attribute) – Sizes of nodes are necessary to know the size of communities in aggregate graphs. Usually this is set to 1 for all nodes, but in specific cases this could be changed.
SurpriseVertexPartition¶
- class louvain.SurpriseVertexPartition(graph, initial_membership=None, weights=None, node_sizes=None)¶
Bases:
MutableVertexPartitionImplements (asymptotic) Surprise.
Notes
The quality function is
\[Q = m D(q \parallel \langle q \rangle)\]where \(m\) is the number of edges,
\[q = \frac{\sum_c m_c}{m},\]is the fraction of internal edges,
\[\langle q \rangle = \frac{\sum_c \binom{n_c}{2}}{\binom{n}{2}}\]is the expected fraction of internal edges, and finally
\[D(x \parallel y) = x \ln \frac{x}{y} + (1 - x) \ln \frac{1 - x}{1 - y}\]is the binary Kullback-Leibler divergence.
For directed graphs we can multiplying the binomials by 2, and this leaves \(\langle q \rangle\) unchanged, so that we can simply use the same formulation. For weighted graphs we can simply count the total internal weight instead of the total number of edges for \(q\), while \(\langle q \rangle\) remains unchanged.
References
- 1
Traag, V. A., Aldecoa, R., & Delvenne, J.-C. (2015). Detecting communities using asymptotical surprise. Physical Review E, 92(2), 022816. 10.1103/PhysRevE.92.022816
- Parameters
graph (
ig.Graph) – Graph to define the partition on.initial_membership (list of int) – Initial membership for the partition. If
Nonethen defaults to a singleton partition.weights (list of double, or edge attribute) – Weights of edges. Can be either an iterable or an edge attribute.
node_sizes (list of int, or vertex attribute) – Sizes of nodes are necessary to know the size of communities in aggregate graphs. Usually this is set to 1 for all nodes, but in specific cases this could be changed.